
DNA is a double-stranded helix but RNA is single-stranded. Both are built using nucleotides (adenine, cytosine, guanine, however, in RNA, thymine is replaced by uracil). MRNA and DNA are different forms of the same language. Instead of students, they are RNA polymerases. In the real world, instead of copying the instructions down as words on a page, it’s copied down (transcribed) as mRNA or messenger RNA. To complete the assignment, students need to first copy down the instructions into their notebooks and then go home to work on it. Let’s imagine DNA as an assignment on a whiteboard.

The transformation occurs by converting DNA to mRNA and then to a protein. Like I mentioned before, DNA needs to be transformed into a protein to be functional. In fact, I have a bracelet that perfectly illustrates this. Make sure that your enzyme only has 1 cut site (in the multiple cloning site) or else your plasmid will become sliced like a loaf of bread and disintegrate. Cut sites are parts along the genome that your chosen restriction enzyme can bind to and cut at. However, as a plasmid designer, the most important property you have to keep in mind is the number of cut sites. Having a variety of restriction sites also makes it more convenient for scientists to choose a perfect one (based on cut sites, availability of enzymes, cost of enzymes, etc.) They enable scientists to insert a piece of DNA without disrupting the rest of the plasmid. Multiple cloning sites are short segments of DNA that contain up to 20 restriction sites. In plasmids engineered for commercial use (for researchers, companies, etc.), restriction sites are often clumped together in multiple cloning sites (MCS). To prevent lab work from being even more expensive than it already is, scientists modify plasmid backbones to optimize for success. But could you imagine if we brought the wilderness into the lab and just hoped for everything to go as intended? Everything is left to chance and rarely goes right. Chances of successful plasmid mutation and infiltration are at an all-time low. However, in nature, it’s a dog-eat-dog world out there. Plasmids are actually naturally found in bacteria and enhance their survival. Now that there’s a gap in the DNA, researchers can insert a gene into that gap and form a recombinant plasmid. Each restriction enzyme has its own unique restriction site. Once found, they go *hiyah* and form a double-stranded break at that site. Wielding their molecular katana swords, they search the genome for a specific complementary 4–8 bp sequence called a restriction site. You can think of restriction enzymes as DNA ninjas with one sole mission. Restriction Sites and Restriction Enzymes Watch this video if you want to learn more about DNA replication. Primase takes the replicated plasmid from 0 → 1, DNA Polymerase takes it from 1 → 100(00…). The primer is like a pull tab for a zipper, enabling DNA Polymerase to bind and synthesize the rest of the nucleotides by zipping down the DNA.
The DNA is unzipped by helicases (a protein) and primase (another protein) attaches a short RNA-chain called a primer.

The origin of replication or ori is a specific region within the plasmid’s DNA sequence that tells the host cell’s replication machinery to “bind here!” and replicate the entire plasmid. Plasmids need to reproduce - make copies of themselves - to survive and live on.
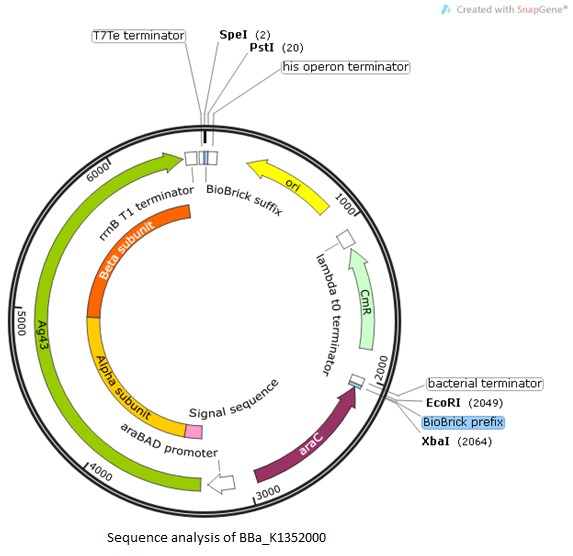
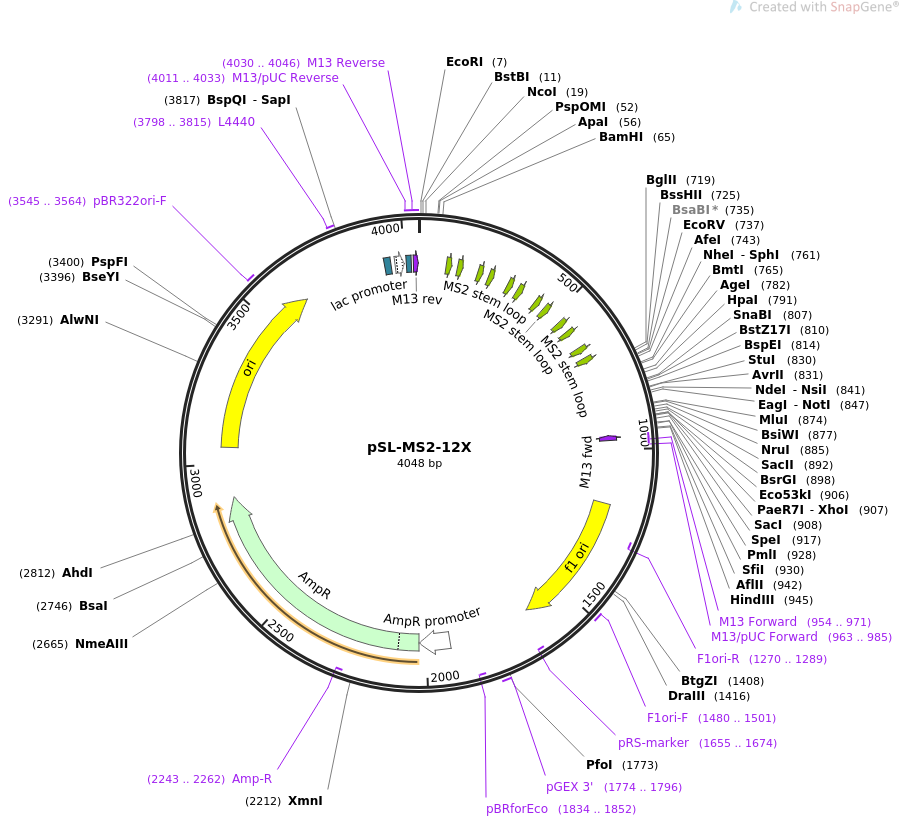
In real life, they’re not colour-coded or labelled, and cells can still identify everything perfectly. Let’s break them all down.įor clarification, everything above is all just DNA (A’s, C’s, G’s, and T’s). Every plasmid has 8 main elements: the origin of replication, restriction sites, a promoter, terminator, inserted gene, antibiotic resistance gene (prokaryotic) or auxotrophic selection marker (eukaryotic) and start & stop sequences for translation.


 0 kommentar(er)
0 kommentar(er)
